
Stories
Gregory Hannon: Tools for Tough Questions

Gregory Hannon (Photo: CRUK Pioneer Award)
Gregory Hannon grew up in western Pennsylvania, where his father worked as a quality inspector at a factory. Hannon had professional aspirations, and assumed he was “destined” to become a physician. “Growing up, I didn’t even know that the job I do now existed,” he explains. As an undergraduate at Case Western Reserve University, Hannon took a job as a lab assistant, and this taste of the research life changed his plans.
Working with structural biologist Joyce Jentoft, Hannon learned to use nuclear magnetic resonance (NMR) spectroscopy to determine the structures of proteins, and wrote one of the first software programs for visualizing protein structures on a personal computer. Hannon stayed at Case Western for his Ph.D., delving into the mysteries of RNA processing in the laboratory of Timothy Nilsen. His research was fueled by an unexpected turn of events: another student in the lab got scooped on the discovery of trans-splicing—a phenomenon in which RNA molecules transcribed from different genes are joined together and translated into a kind of fusion protein.
Although others had demonstrated the existence of trans-spliced RNAs, the production mechanism of these oddball molecules remained unknown. So Hannon and his colleagues examined trans-splicing in Ascaris lumbricoides, a giant parasitic roundworm with eggs that could be easily collected and induced to develop synchronously—helpful properties for gathering sufficient RNA material to visualize and study.
By investigating trans-splicing in cell-free extracts from developing embryos, Hannon was able to decipher the details of the reactions that append a specific “spliced leader” sequence of nucleotides to an RNA molecule, thus marking it for trans-splicing. Further research revealed the identities of the molecular labels that enable two strands of disparate RNA to associate and recruit proteins that splice them together. This work “added new twists to our understanding of RNA metabolism,” Hannon says. Today, researchers are investigating the potential of trans-splicing to correct disease-causing mutations.
Hannon switched gears as a postdoc, moving to Cold Spring Harbor Laboratory and working with yeast geneticist David Beach to study the activities of cyclin-dependent kinases, key cell cycle regulators whose mutation can lead to cancer. Hannon and Beach discovered a tumor suppressor gene, p15, whose product inhibits cyclin-dependent kinases. When it functions properly, p15 can respond to external signals by halting cells in an early phase of the cell cycle, preventing further growth and cell division. Hannon contributed to the discovery of two related cyclin-dependent kinase inhibitors, p16 and p21. Together these represented a new class of cell cycle regulatory proteins, which have turned out to play major roles as tumor suppressors.
After joining the faculty of Cold Spring Harbor Laboratory in 1994, Hannon continued to explore the underpinnings of cell cycle control. By the time he was named a Rita Allen Foundation Scholar in 2000, Hannon had shifted his focus to developing genetic tools, driven by a desire to go deeper into biological questions than biochemistry had allowed him to. “If there’s one theme, it’s that we don’t really have a single field,” he says of his team’s work.
Hannon studied the mechanisms of RNA interference—a process by which a double-stranded RNA molecule can block a complementary messenger RNA from being translated into a protein, effectively “silencing” the corresponding gene and allowing researchers to deduce the normal function of the silenced gene. Hannon’s group successfully targeted specific transcripts for silencing in fruit fly cells, demonstrating that the silenced transcripts were enzymatically degraded.
Later, they characterized the enzymes Dicer and Argonaute, central players in the silencing process, and developed a method to stably knock down gene expression in mammalian cells. By 2002, so many scientists were exploring various aspects and applications of RNA interference that Science Magazine declared small RNAs “Breakthrough of the Year.” (Andrew Fire, a 1989 Rita Allen Foundation Scholar, shared the 2006 Nobel Prize in Physiology or Medicine for the discovery of RNA interference.)
“As scientists, we produce science—we produce results, we produce insights. But I think it’s much more important that we produce scientists.”
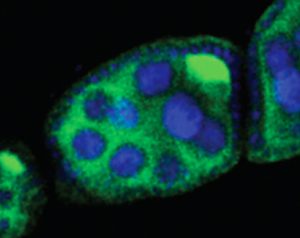
A developing fruit fly egg chamber, showing “nurse cells” that support the egg (blue) and a transposon (bright green), which will lead to sterility in the fly that develops from this egg. Rita Allen Foundation Scholar Gregory Hannon studies the Piwi-interacting RNA pathway, which protects the genomes of gamete-producing cells from disruptive transposons.
(Image: Hannon Lab/CSHL)
Hannon went on to make key advances in understanding the Piwi-interacting RNA pathway, which is vital for fertility in all animals, as it protects the genomes of gamete-producing cells from disruptive transposons. In 2014, Hannon moved his lab to the Cancer Research UK Cambridge Institute.
Hannon has served on the Rita Allen Foundation’s Scientific Advisory Committee since 2009. Here, he describes his renewed focus on tumor biology, his drive to develop new research technologies, and the joys of training and supporting young scientists.
Where are you going next in your research?
In our cancer research, we’re trying to understand tumors in 3-D, or better yet, in 4-D. We want to characterize a lesion as a whole: Which cells
are there, what are the cells doing, and how are they communicating with each other? How does heterogeneity within tumors impact both disease initiation and progression, and especially treatment response? Now that we’ve moved to a place that is very cancer-focused, we hope to use what we’ve learned to have a direct impact on patients.
In terms of basic biology, we want to better understand the mechanistic details of the Piwi- interacting RNA pathway—a deeply conserved process that is absolutely critical for defending germ cell genomes. It’s a very complicated small RNA-based system that operates in every animal and is essential for fertility.
We want to understand how the system is put together, down to the level of protein structures and interactions. We’ve had lots of surprises so far, including RNA acting as a means of epigenetic inheritance. I think there will be a lot more surprises as we learn more about the mechanistic details, and I think those insights will flow into other related fields—less specialized than these very specific germ cell phenomena.
What are the greatest challenges in your work?
The big challenge is that almost everything in science is technology-related. If the technology is available to ask an important question, it gets asked. We tend to do things that aren’t quite possible with toolkits that are out there, so we’re always running up against technical barriers to answering questions that are deeply biological.
In any thread of work in my lab over the last couple of decades, there are always key points where we’ve either made or adopted some cutting-edge approach that transformed what we could do. There are half a dozen of these projects in the lab right now. For instance, we’re trying to work with very small bits of clinical sample to determine how we can predict whether early breast cancer will progress or not.
There are challenges around trying to understand the biology of very rare dormant cell populations, where you’ve got to pick through a whole tissue to find one cell and figure out what it’s doing—which neighbors it’s talking to and how. These kinds of problems are at the edge of what is currently possible. So we spend most of our intellectual effort thinking about solutions to these technical issues.
What do you see as the most significant impacts of your work?
We’ve tried to develop really good tools, get them into people’s hands, and hope that, by having them deployed broadly, we can have an impact on our basic understanding of science, but also have a clinical impact that is beyond what we could do as an individual lab.
The other way to magnify your impact is by training people. As scientists, we produce science—we produce results, we produce insights. But I think it’s much more important that we produce scientists. When I look at the people who have been trainees in the lab, and at where they’ve ended up in their careers, it amazes me constantly. It’s great to hear from them when they get their first job, have their first child or publish their first paper from their own lab. They still want advice on things, and it reminds me that I’m building this science family—it’s like having children. One of the most important things I’ve learned is that we are teachers before anything else.