
Stories
Tom Maniatis: Mastering Methods and Exploring Molecular Mechanisms
Tom Maniatis launched his scientific career by experimenting with early ultraviolet lasers to scrutinize the development of chicken embryos. This undergraduate research experience, at the University of Colorado, Boulder, “taught me the importance of bringing technological developments to important biological problems,” Maniatis recalls.

Tom Maniatis
(Photo: Columbia University Medical Center)
Devising and applying new technologies have since become hallmarks of Maniatis’ work, from pioneering protocols for identifying, isolating and cloning genes in the late 1970s, to more recent investigations of the nervous system using mouse models and stem cells.
As a Ph.D. student with Leonard Lerman at Vanderbilt University, Maniatis established new methods for studying the structure of compact DNA using X-ray scattering, with relevance for understanding DNA dynamics in both chromosomes and viruses. These investigations “required designing and building instruments, and using some of the earliest computer algorithms to interpret the diffraction data,” Maniatis notes.
Fascinated by the emerging capabilities of molecular biology to reveal the intricate forms and functions of genetic material, Maniatis moved to Harvard University for a postdoctoral fellowship. He worked with Mark Ptashne (now at Memorial Sloan Kettering Cancer Center) to characterize gene regulation in bacteriophage lambda, a virus that infects E. coli bacteria and has become both a model system for understanding genetics and a key tool for genetic engineering. Maniatis and Ptashne traveled to Fred Sanger’s laboratory in Cambridge, England, to make use of Sanger’s still-in-progress DNA sequencing techniques, for which Sanger later received a Nobel Prize in Chemistry. The characterization of the protein-DNA interaction in phage lambda set the stage for understanding the “genetic switch” in the life cycle of the phage—between dormancy, or lysogeny, and lysis, in which the phage kills the host cell and releases newly assembled viral particles.
Maniatis and his colleagues then began forging a new approach for studying genes in eukaryotic organisms (plants, animals and fungi)—a process for cloning complementary DNAs (cDNAs, the functional versions of genes produced from messenger RNA precursors) and using these cloned cDNAs to isolate the corresponding genes from genomic DNA libraries. Maniatis was unable to continue this work at Harvard, however, due to a moratorium on recombinant DNA research in the city of Cambridge, Massachusetts.
Fortunately, Maniatis was offered faculty positions at Cold Spring Harbor Laboratory and then at Caltech, where he was a Rita Allen Foundation Scholar from 1978 to 1979. This was a fruitful period for his research, as he completed the development of the first full-length cDNA clone (beta-globin) with his collaborators Fotis Kafatos and Argiris Efstratiadis at Cold Spring Harbor. Subsequently, Maniatis and his team at Caltech developed the first complete genomic DNA libraries—collections of cloned copies of all the genomic DNA in Drosophila (fruit fly), rabbit and human cells. “The Drosophila and human libraries were widely used by the Drosophila developmental biology and human genomics communities for many years to isolate and study individual genes,” Maniatis says.
Using the human genomic DNA library, the Maniatis group was the first to clone human genes—delta-globin and beta-globin, which encode two subunits of hemoglobin, the vital oxygen-carrying protein found in red blood cells. Maniatis’ group went on to characterize the full structure of the human globin gene cluster and map genetic mutations in the globin genes responsible for forms of an inherited disease called thalassemia.
The city of Cambridge eventually lifted its ban on recombinant DNA and established new research regulations. In 1980, Maniatis returned to Harvard, where he helped to shape the University’s nascent Department of Molecular and Cell Biology. Around the same time, he also embarked on a project that would lead to perhaps his most enduring contribution to science: Maniatis coauthored Molecular Cloning: A Laboratory Manual, which was first published in 1982 and has since become ubiquitous in laboratories throughout the world—a trove of protocols and tips for investigating genetics and cell biology. In 2012 Maniatis received the Lasker~Koshland Award for Special Achievement in Medical Science; the award announcement cited the transformative impact of the manual and praised Maniatis for “fundamental discoveries concerning the nature of genes.”
“When we developed cDNA and genomic cloning methods, we clearly recognized the potential impact of these methods on eukaryotic molecular biology. Less obvious were the enormous impact that the molecular cloning manual would have on the propagation of the technology worldwide, and its impact on biomedical science.”
Maniatis has continued to make use of ever-advancing molecular tools to study the regulation of immune responses. His lab has also made fundamental contributions to understanding the mechanisms of pre-mRNA splicing.
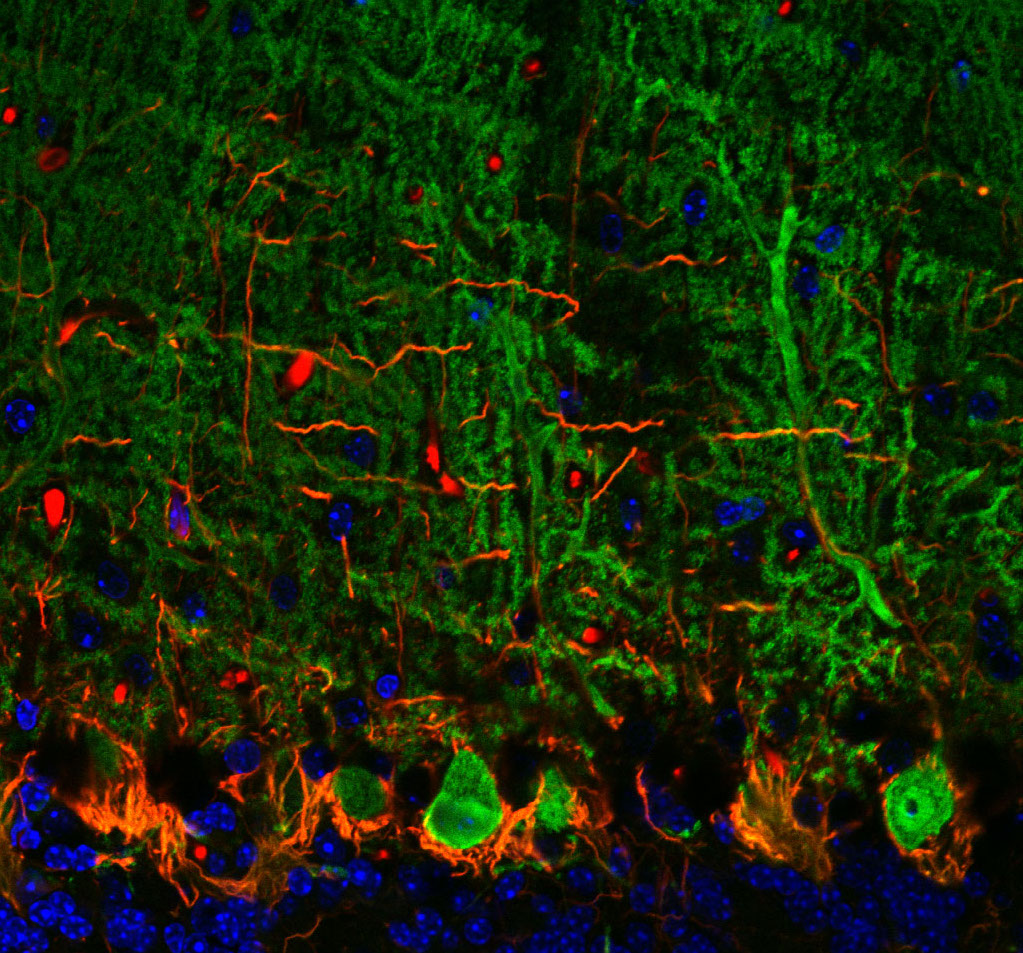
Rita Allen Foundation Scholar Tom Maniatis studies molecular neuroscience, including the disease mechanisms of amyotrophic lateral sclerosis (ALS). ALS involves degeneration of Purkinje neurons, which are among the largest brain neurons and form abundant dendritic spines. Purkinje neurons (shown here in green) function in motor coordination in the brain’s cerebellum.
(Image: Wikimedia, Sbrandner)
He moved to Columbia University in 2009, and during the past two decades he has branched out into neuroscience, pursuing the function of protocadherins—a large family of cell surface proteins—in brain wiring. In addition, prompted by his sister’s death from amyotrophic lateral sclerosis, or ALS, he now investigates the underpinnings of this neurodegenerative disease.
Here, Maniatis shares his thoughts on the far-ranging influence of molecular cloning methods and reflects on the next frontiers of his research in neuroscience.
When you helped to develop methods and write the Molecular Cloning manual, what did you think the impact of this work would be? Has anything surprised you about the development of molecular biology and recombinant DNA technology?
When we developed cDNA and genomic cloning methods, we clearly recognized the potential impact of these methods on eukaryotic molecular biology. Less obvious were the enormous impact that the molecular cloning manual would have on the propagation of the technology worldwide, and its impact on biomedical science.
I was a Rita Allen Scholar at Caltech when I was asked by Jim Watson [best known for helping to discover the structure of DNA; also an emeritus member of the Rita Allen Foundation’s Scientific Advisory Committee] to teach the first “Molecular Cloning” course at Cold Spring Harbor. Ed Fritsch, then a postdoc in my lab, and I assembled all of the cloning and analysis protocols from my lab in a loose-leaf notebook and prepared reagents for the course.
The cloning course was a great success, and at the end of the course Jim asked me to turn the course notebook into what became the Molecular Cloning manual. He also convinced Joe Sambrook, at that time the scientific director at Cold Spring Harbor, to join the effort, and I convinced Ed Fritsch to join. The hardest part was integrating the necessary biological background with the protocols in such a way that novices could learn to troubleshoot. Others have told us that this was, in fact, the basis for the success of the manual. It allowed naive investigators who did not have access to nearby experts to successfully apply the then-new technology to their biological problems. This, in conjunction with the timing of publication (early in the application of the technology), led to an enormous impact, internationally, in nearly every aspect of biomedical research. I believe that only Jim Watson fully understood the potential impact of the manual.
When you think about the future of your research and your fields of study, what possibilities are most exciting to you?
The development of recombinant DNA methods in the late 1970s was just the beginning of rapid and dramatic technological advances, ranging from the development of sophisticated tools for genome manipulation (including the recent development of CRISPR technology), to optogenetics and high-throughput DNA and RNA sequencing, to single-cell and single-molecule technologies. These technologies are profoundly advancing our understanding of the brain.
My lab is currently focused on two projects in the area of molecular neuroscience. The first is directed toward understanding the expression and function of the clustered protocadherin genes, which we discovered more than 15 years ago. New technologies have made it possible to probe deeply into the molecular mechanisms involved in the generation and function of single-cell diversity in the brain, the structure and function of protocadherin proteins, and, most recently, the function of protocadherins in the development of brain circuitry in vivo. Massive whole-exome [expressed genes] or genome sequencing studies of individuals with autism have led to the identification of DNA sequence variants in the protocadherin gene cluster, and these sequence changes occur in sequences which we know to be essential for protocadherin function. Thus we are poised to understand not only the function of protocadherin genes in brain wiring, but also their possible role in neurological diseases.
Our work in ALS has been similarly impacted, with the identification of close to 20 new ALS genes during the past decade based on next-generation sequencing technology, and progress in understanding ALS disease mechanisms. These important advances in genetics have identified new pathways and potential targets for drug development. Our particular focus is currently on ALS genes that function at the interface between innate immunity, inflammation and autophagy [the destruction of cells]. I think it is fair to say that more has been learned about ALS genetics and disease mechanisms in the past 10 years than in the preceding 100 years.